We aimed to find out STK31 as a cancer-testis (CT) gene and to find its potential medical price, regulatory mechanisms, and gene neighborhood in pancreatic most cancers (PC).
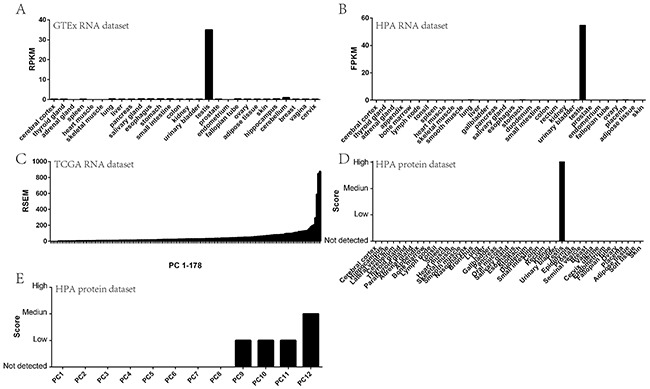
Gene expression information had been generated from common organ samples and pancreatic most cancers samples from three public databases. STK31 expression patterns in common and PC tissues had been acknowledged, and we explored its regulatory mechanisms.
Gene ontology (GO) and pathway analyses of STK31-related genes had been carried out and an STK31 protein-protein interaction (PPI) neighborhood was constructed.
STK31 was confirmed as a CT gene in PC and its expression was significantly elevated in victims with new neoplasm in distinction with victims with out new neoplasm (P = 0.046) and in extra superior pathologic ranges than in earlier ranges (P = 0.002); methylation diploma correlated negatively with STK31 expression.
In full, 757 STK31-related genes had been acknowledged, and had been significantly enriched in terms of polymorphisms and varied splicings. The PPI neighborhood predicted that STK31 was bodily associated to the PIWI (initially P-element Induced WImpy testis in Drosophila) and Tudor households.

Newest genomic analyses in Drosophila and mammals of inter-chromosomal retroposition have revealed that in evolution the retroposed genes that current male-biased expression generally tend to go away the X chromosome and go for autosomal positions.
Such a phenomenon may be a technique of fundamental, genomic and evolutionary relevance. It contributed to the sudden overrepresentation of male-biased genes on the autosomes currently observed in microarray expression experiments.
On this paper, we report our genomic analysis of within-chromosomal retroposition in Drosophila melanogaster, and consider it with the beforehand acknowledged pattern of the between-chromosomal retroposition.
We uncover {{that a}} surfeit of autosomal retroposed genes originated from parental genes positioned on the similar chromosome, in distinction to the X chromosome by which solely few genes retroposed in cis. Such an autosomal proximity influence implicates a job of the mutation course of for retroposition in figuring out chromosomal areas of autosome-derived retroposed genes.
Furthermore, this phenomenon helps the hypothesis that pure alternative favors the retroposition of genes out of the X chromosome. Analyses of an enormous expression database for D. melanogaster genes revealed that the overwhelming majority of the X-derived autosomal retroposed genes had developed testis expression options, in keeping with completely different earlier genomic analyses.